
Home
Mission
Overview of Project
Project Staff
Sponsors
Achievements
Checking, Illustrations
Upcoming Activities
Id and Species Lists
Protea Information
Protea Gallery
Growing Proteas
Interim Dist. Maps
Publications
Afrikaanse Inligting
![]()
WORLDMAP
WORLDMAP and The Protea Atlas Project
The Protea Atlas Project was initiated in 1991 to map the distributions of species of the Protea Family for a variety of purposes. Astonishingly, we do not know where many of our protea species occur. Thus the location of populations of some widespread species, known to thousands of South Africans, are unknown to conservationists for the simple reason that "everyone knows that they occur there". At the other extreme are some extremely rare species which could do with more public attention, such as Serruria aemula, which is unknowingly passed twice a day by thousands of motorists as they commute to and from Cape Town. Knowledge of the former type is required so that we can effectively plan and assess our conservation plans. Knowledge of the latter type is required so that some highly threatened species do not accidentally become extinct. Both aspects require that the general public be made aware of their surroundings, be educated about the veld and get to know their plants. The Protea Atlas Project attempts to initiate this awareness.
But distribution data can be used for many other things: for planning conservation management, for locating genetic resources, for assessing tourist potential of particular areas at particular times of the year, for education of local communities, and the list is endless. But it all depends on the data being used and communicated.
So I was greatly excited when the Natural History Museum of London granted us permission to use their programme WORLDMAP as an interface between data and atlassers. Obviously, we cannot make all our data available to everybody: some sites are sensitive and access should be limited (other sites are in dire need of constant attention, but we attend to these in other ways); some species are prone to casual exploitation and require a degree of confidentiality; some people cannot be entrusted with too-detailed data (fortunately proteas are not as sensitive as cycads, succulents or bulbs); and, there are just too much data coming in to be handled by many atlassers PC's. So we settled on a 12km X 12km grid square system for three reasons: I had used it for my doctorate and so herbarium data were available for Proteaceae; the cells can be obtained by dividing a South Africa 1:50 000 map into four equal units; and, it reasonably approximates the 10km X 10km grid square system used in the UK and some other regions. We are also investigating a 25km X 25km grid square system for southern Africa (which is the same as the 1:50 000 series maps - this should be available early in 1996).
More excitingly, atlassers can now be on the cutting edge of science. This programme is the state of the art in reserve location theory, amalgamating theoretical advances from the United Kingdom, South Africa and Australia. Not only will atlassers have access to the Protea Atlas Project data, but they can play with designing efficient reserve systems for protecting our flora. Because we will be maintaining it as a current data base, with updates every year, atlassers can assess for themselves how our data are affecting conservation decisions. Of course, there is no reason why you should stop with the Protea Family - it is easy enough to enter your own data on any plant or animal group in which you are interested. But that is another atlassing project! The condition of use is that if you use WORLDMAP for your own data you must send a copy of your data to the Natural History Museum in London. A maximum of about 500 taxa can be stored given 4 Mb of free RAM.
General requirements are similar to those for running Microsoft_WINDOWS version 3.1. The exception is that there is no support in WORLDMAP 3 for the VGA graphics mode, because the 640 x 480 pixel resolution is inadequate for map graphics. You will need:
- IBM-compatible computer with a DOS operating system and mouse
- 80386 or 80486 processor
- 4 Mb or more RAM (memory)
- Super-VGA graphics card (preferably 1 Mb for 256 colours) with a colour monitor.
To obtain WORLDMAP with the latest Protea Atlas Data, please submit a single stiffy disc to the Protea Atlas Project (with your name and address on the label) and we will send you the software and data by return of mail. Alternatively, e-mail me and I will send you the programmes via the ether. It will help if you tell me your computer type (386 or 486) and your graphics mode (trident, etc.).
WORLDMAP is controlled by clicking one of the mouse buttons while the graphics cursor is positioned over the required 'button' on the screen.
The data base, which can be updated in editing mode, consists of three parts:
- The Species Name, which is the genus and species name (limited to x characters).
- The Clade Code, which is a alpha-numerical string which summarises the cladogram for the species under consideration. This is handy when one wants to restrict analysis (say species richness or reserve selection) to a single subfamily, genus, subgenus, or section, rather than the entire data base. Thus the first branching is coded as group 1 and group 2. The first subgroup in group 2 is group 21, etc. In analyses the species are listed in the sequence of their Clade Code.
- The Map (or Distributional) Data, which is displayed in editing mode as a map for the species.
Two levels of data are possible: for the Protea Atlas Project we use herbarium data as level 1 (grey spot) and "atlassed" as level 2 (white spot). Analyses can be applied to both levels (1 & 2) or to level 2 alone. Data can be amended by merely clicking the mouse on the relevant square on the map.
Three types of printed output can be generated from the above data base in editing mode:
- A list of the species and their clade codes;
- A list of species records for each grid cell; and
- A list of grid cells occupied by each species.
When one is not in editing mode the map can be used for analysis. The simplest is to point and click on a map square and a list of all species in that square will appear on the right hand side of the screen next to the map. Clicking the mouse again replaces the species list with a diagram showing the position of those species on the cladogram.
By clicking on the Right Hand Side buttons one can look at various measures of biodiversity:
- "Taxon richness" will display the number of species in each grid square
- "Char richness" will display the proportion of the cladogram represented, . and,
- "Endemism" will display the degree to which the squares contain very rare species.
In each case the squares are shown on a colour-coded scale from blues (few species or low scores) to reds (many species or high scores). By pointing and clicking on squares of interest on the map one can get their species listing and cladograms.
However, the real fun of WORLDMAP is determining conservation options and priorities. This can be done in many ways. The easiest is to select the "Auto select" button and answer a few queries so that it toggles from "-" to "+" and then hit one of the above buttons. This will determine the minimum set of grid squares are required to conserve all the species in as many reserves as you specified. Not only do you see the sequence in which reserves are selected, but the most important reserves are coded red through to the least important in blue.
Far more fun is to use the "Exclude Areas" button. The method is simple. Select the diversity measure you want (e.g. Taxon richness), and then hit the "Exclude Areas" button. The hottest areas of biodiversity are shown in red and by clicking on the square you "add" that square to your nature reserve list. You can also add areas which have nature reserves and areas where you will like to see made into nature reserves. You can thus play with how you would design a nature reserve ("excluded areas") system. You can test the effectiveness of your system by clicking on OK and now the map will display the biodiversity of those species not included in your nature reserve system. Clicking on the squares of the map will again produce the species lists and cladograms, but with the species which are "conserved" and those which are not in different colours. By toggling between the two buttons you can add to or refine your nature reserve system, or determine which nature reserves are superfluous.
If in the "Options" button you have selected "Results file +" (by clicking your mouse on the button before you began) you can have a printout of all your steps, as shown below: Thus step one "conserved" 23 species in area 459, but adding area 536 (with no species) at step two achieved nothing. In step three, area 476 had two species of which one was added to the reserve system (the other was already included in area 459). By step 18 we had "conserved" 98 species in 18 reserves, some of which (areas 536 and 771) are clearly superfluous existing nature reserves (for proteas!).
(Thu Jul 28 16:52)
Big-grid WORLDMAP - Version 3.18
Single-step priority-areas analysis: Proteaceae in the Cape Flora.
=============================================
Diversity measure: (Ia) ENDEMISM WEIGHT.
(data levels 1&2)
| Choices | Taxa | Diversity | ||||
| step | area | abs | inc | cum | inc % | cum % |
| 1 | 459 | 23 | 23 | 23 | 1.25 | 1.25 |
| 2 | 536 | 0 | 0 | 23 | 0.00 | 1.25 |
| 3 | 476 | 2 | 1 | 24 | 0.27 | 1.53 |
| -- | -- | -- | -- | -- | -- | -- |
| 16 | 744 | 28 | 20 | 92 | 6.14 13.52 | 329 |
| 17 | 764 | 7 | 6 | 98 | 0.96 14.48 | 503 |
| 18 | 771 | 0 | 0 | 98 | 0.00 14.48 | 856 |
By clicking on the "Subgroup" button you can select only a particular clade (e.g. 2 (Pincushion allies) or 223411 (Sandveld Conebushes) for analysis. This does not affect the data base, only the above analyses.
The "Options" button allows you to set your system needs. For instance, the "Data level 1&2" button allows you to analyse the entire data base (levels 1&2) or just level 2, or, using other buttons, you can reset some of the buttons on the main menu, or adjust colours for biodiversity scores on the maps.
Already some interesting data have turned up. For instance:
The Kogelberg does not appear to be the centre of the Cape Floral Kingdom. More species are found in grid squares near Villiersdorp. This is not a sampling problem - early atlas data mirrored the herbarium data, and at least one additional "undescribed" protea species (in the genus Serruria) is known from the area.
Many species which were once collected extensively on the Cape Flats are now apparently extinct there. This requires that additional conservation areas on the northern and eastern borders of Cape Town be established. Should this not be possible, several species will become extinct, and with them several non-protea threatened species.
Many localised species have turned out to be more widespread than thought - fully one-third of all proteas have had their distribution ranges extended and another third have had gaps in distributional data filled in.
Several species are very poorly atlassed - far more so than can be expected from historical data - what has happened to these species? Have they become extinct over much of their historical distribution range, or have collectors/botanists changed their hiking habits? Why has this pattern/trend not been picked up before?

The above map shows species richness of proteas (i.e. the Family Proteaceae) in the Cape Flora. Note that the traditional area of species richness at Kogelberg State Forest has been usurped by the western Riviersonderend near Villiersdorp! (Red implies many species - Dark Blue implies few species).
Have a look at more on Species Richness for Pincushions and Spiderheads and the latest Species Richness map for all the Cape Flora Proteaceae at November 2001.
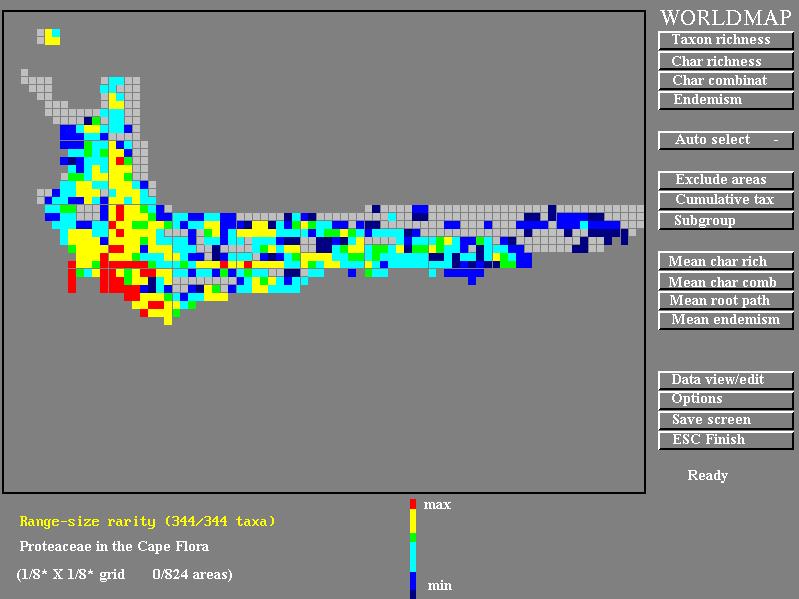
The WORLDMAP screen. The feature selected is "Endemism" and the distribution of rare species can be seen to be towards the west of the region. The buttons for selecting options are shown on the right.
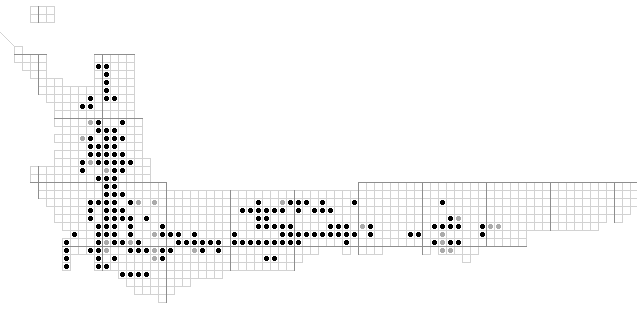
The WORLDMAP editing screen. The species shown is Protea nitida and the atlas data (option two) are shown in black) and the herbarium data which have not yet been verified (option one) are shown in grey. Editing the distribution map can be accomplished by clicking on the square which toggles it to the next of option 1, option 2 or option 0 (absent).
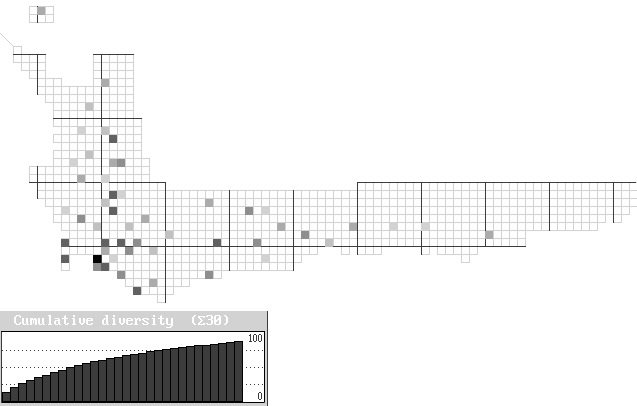
The end result of an automatic analysis of the minimum number of squares required to preserve all the proteas in a conservation network. This analysis was done on all records (atlas and herbarium), with no pre-existing reserves, so that each species is represented in at least one square. Only the first 30 grid squares are shown on the graph, which shows that over 90% of species are represented in the first 25 squares. Conserving the last 10 per cent of species requires over half the total grid squares (the first 10% was conserved in a single grid square). Checking of the output in the results.out file will show which squares are irreplaceable (their unavailability will either result in the extinction of a species or the addition of more than one additional grid square to the reserve system) and which ones are flexible (i.e. can be substituted by another grid square[s]). For more details on iterative procedures see Rebelo (1994).
(Published in PlantLife 15, Sept 96).
You may download Protea Atlas datafiles for Worldmap by clicking on the below links. Make sure that that the downloaded file is placed in the \Worldmap\Cape directory. Each download includes the Worldmap locality file (Worldmap.loc) which you should also copy to your \Worldmap\Cape directory. All downloads contain exactly the same locality file.
Pre Protea Atlas Data (37kb)
Protea Atlas Data 1996 (41kb)
Protea Atlas Data 2000 (42kb)
Protea Atlas Data 2001 (42kb)
All the Protea Atlas data for Worldmap (All of the above) (92kb)
Back History