
Home
Mission
Overview of Project
Project Staff
Sponsors
Achievements
Checking, Illustrations
Upcoming Activities
Id and Species Lists
Protea Information
Protea Gallery
Growing Proteas
Interim Dist. Maps
Publications
Afrikaanse Inligting
![]()
A Phylogeny of Proteas
Gail Reeves, Jodrell Laboratory, Kew
The primary aim of my PhD research is to understand the evolutionary relationships among species of the genus Protea. The genus currently comprises approximately 114 species, which are found throughout South Africa (69 species are endemic to the Cape Region alone) and tropical Africa.
Species of Sugarbushes exhibit a wide array of diversity of morphology and ecological specialisation. This is reflected in their preferred choice of soil type, fire survival (e.g. reseeders vs resprouters) and pollination biology (e.g. by birds, rodents, etc.). Largely as a result of your dedication as atlassers Protea represents one of the most extensively documented groups of the flowering plants from an ecological standpoint. Whilst this wealth of information serves as a excellent tool in conservation management, students like myself can also use this data to address some fascinating questions posed by evolutionary biology.
As you know from your - no doubt well-thumbed - SASOL "bible", the genus Protea is currently split into various sections. These compartments are indispensable as they aid identification based upon floral and vegetative characters that are easily observed with the naked eye.
However, the way in which the species have been grouped may not necessarily reflect how they are actually related to one another. In other words, species may share characteristics that have led taxonomists to group them together, but these groupings may be "artificial" in that they do not represent groups of closely related species. Another issue that is difficult to address using whole plant characteristics is how the groupings in the genus are themselves related to each other. For example, we may hypothesise that the Eastern and Western Ground Sugarbushes are each other’s closest relatives but how are the Bearded or Spoon-bract Sugarbushes related to these?
The aim of my study is to reconstruct the Family Tree for all the species of Protea. A phylogeny is a representation of how taxa (species, subspecies, and higher groups – sections, subsections, subgenera, subfamilies, etc.) are related to one another. The figure below shows some very preliminary results that I have so far. Obviously the sampling is incomplete at this stage, but already we can see a pattern emerging. The supposedly more "primitive" species comprising the Grassland and Shavingbrush Sugarbushes form a distinct group with the remaining, more "derived" species, forming a second. Note though that, at this stage, Pr dracomontana is more closely related to the Shavingbrush than the Grassland Sugarbushes!
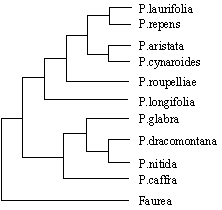 |
Grassland and Shaving-brush Sugarbushes |
Once the phylogeny is complete we then plan to use this framework to look at how the present day distribution of morphological and ecological characteristics have evolved. For example, did the ability to resprout after fire evolve once during the evolution of the genus, or did it evolve several times in response to similar ecological pressures in separate geographical locations. Also, did the axillary flowerhead displayed by the rodent-pollinated Sugarbushes evolve just once or independently in several lineages? These are just some examples of the questions that an accurate phylogeny will help to answer.
Using DNA sequences to build robust phylogenies represents the most modern approach in systematics. The most obvious advantage to using DNA sequences rather than morphological characters to build a phylogeny is that it is possible to collect several thousand DNA characters from each taxon (as each DNA base represents one character). The most morphological characters you can reasonably expect to collect for a group ranges between 30 to 50. The assumption is that the more characters (i.e. the more evidence) that can be collected for a group the more accurate the resulting phylogeny will be.
So here lies my justification for spending three months in South Africa with Tony et al. hunting for Proteas. My aim was to collect a few grams of leaf material, from a representative of each species, for later DNA analysis in the molecular systematics laboratory at Kew Gardens. (I can tell you that I am being punished for having such a wonderful time collecting plants in the field in SA: I have spent the last month here in the lab painstakingly grinding up Protea leaves to extract their DNA. Once completely dried out I can promise you that they are not the easiest things to grind {"liquidizing" them destroys the DNA}).
The topic of DNA extraction leads me to one of the questions that Austen Williams set for me in the last newsletter:
"How is the DNA extracted from plant cells?"
The DNA extraction protocol we currently use at Kew Gardens isolates total genomic DNA from the plant, that is all three (nuclear, chloroplast and mitochondrial) genomes. The plant cells are broken open by simply grinding leaf or flower material in a pestle and mortar with a buffer solution. The buffer protects the DNA in solution once it has been released from the cells. The resulting extract is then centrifuged - this separates the DNA from all the debris (cell membranes, cell walls and other contents of the cell). At this point there are several purification steps and finally the DNA extract is suspended in a buffer solution. In this form the DNA can be stored at -80° C for many years. Indeed the "DNA Bank" at Kew now holds DNA extracts from more than 7000 plant species.
Next question "How is the DNA read?"
To build a phylogeny from DNA sequences it is only necessary to target a small number (usually only two or three) gene regions from the thousands of genes encoded by a plants entire genome. So, in actual fact we only "read" very short stretches of DNA at any one time. These sections of DNA are targeted using a process called the polymerase chain reaction (PCR). PCR allows one to target a single gene and amplify it several thousand times until there are many copies of it. In order to actually ‘read’ the DNA sequence we need to label each of the nucleotides in some way so we can detect them visually. There are four fluorescent labels, a yellow one which attaches to the guanine (G) nucleotide, red for thymine (T) , green for adenine (A) and blue for cytosine (C). All the labelled nucelotides are then separated from each another by taking advantage of their electrical charge. The whole cocktail is loaded onto a very thin gel that is poured between two glass plates. An electric current is then applied to this gel and the genes with labelled nucleotides are pulled through the gel past the "DNA Automated Sequencer". The Sequencer uses a laser to excite the labels as the DNA goes past and records the composition and sequence of the nucleotides and saves it as a computer file. Ultimately our final product is simply a precise sequence of the DNA "letters" A, C, G and T, stored electronically for our convenience.
It is the exact order of these nucleotides that is unique to any individual. The same region of DNA is sequenced from each plant and then the ultimate task is to compare the number and composition of nucleotides of each DNA sequence from one plant to the next. We are then able to form hypotheses about how species may be related to one another by using the assumption that species will share more of the same characters with their close relatives, than they will with more distantly related taxa. We have chosen a gene which is being sequenced across all plant families.
A more difficult question:
"On what basis is speciation decided, i.e. must there be a difference in
chromosomes?"
The short answer to this is, no, there doesn’t have to be a difference in
chromosomes, but differences in chromosome number can lead to reproductive isolation
barriers between individuals, meaning that they can no longer interbreed. The Biological
Species Concept describes a species as a "population or series of populations of
freely interbreeding organisms, reproductively isolated from other such populations."
However, this definition of a species we know does not apply to many plant species which
will readily hybridize with each other in the wild (e.g.
Pr longifolia is a promiscuous species that readily forms hybrids with other
species such as Pr eximia). There are rules of thumb about how different the
sequences have to be for species or generic level status, but this is confounded by how
old the taxon is and how recently its elements have been isolated.
One of the burning questions in botany/ecology is simply: "Why are there so many flowering plant species in southern Africa, and fynbos in particular?" Most hypotheses in the current literature point towards the role of fire, and adaptation of species to the different soil types that are characteristic of fynbos landscapes. This suggests that ecological factors may have played a significant part in the evolution of so many plant species, in the Cape Region especially. But alternatively, the Cape Flora may merely represent a set of nutrient-poor islands in a sea of nutrient rich geologies – in which case evolution may be due to successive colonizations as climates have changed in the past. The purpose of my study is to construct a good Family Tree for Protea and then use this to uncover how and why the group evolved into the stunning assemblage we witness today.
Last question! "What are you going to do with 500 angry atlassers if the many lookalikes turn out to be the same species?"
I’m sure that the phylogeny will indicate to us that many Protea species are extremely closely related, especially among those species from the Cape Region. However, I doubt that this will lead to any formal taxonomic revision of species. Don’t worry, all those hours you have put in learning your Protea species will not have gone to waste!!
Before I go, I’d like to say a big thank you to all those atlassers who helped me enormously while I was in RSA last year (you know who you are!). Both for helping me in the field and providing a bed on those occasions when we strayed further from home. Of course, an extra big Thank You to Tony and Val for everything. Will you have me back later this year?
So, that’s about it for this instalment. I’ll keep you up to date with the phylogeny as it takes shape, so watch this space! In the meantime, I’m missing SA loads, so say a collective hello to the proteas from me and spare a thought for those in the Northern Hemisphere, suffering here in the cold! Until next time…
Gail